|
Summary.
The interest of the Sakmar Laboratory in AIDS-related research dates approximately to 1996 with the ground breaking discovery that the HIV-1 virus requires a docking site to enter the interior of CD4+ immune cells. HIV-1 virus particles bind first to CD4, and then to a second receptor, called a co-receptor, to facilitate membrane the fusion between the HIV-1 membrane and the cell membrane, which permits the HIV-1 RNA genome to enter the target cell. HIV-1 co-receptors include heptahelical GPCRs, CCR5 or CXCR4, depending on the viral strain. CCR5 and CXCR4 are members of an important class of GPCRs called chemokine receptors that mediate directed cell migration under normal circumstances. Chemokine receptors comprise a family of about 20 distinct receptors in humans and play key roles in development and in inflammatory responses. Chemokine receptors, especially CXCR4, also play a key role in the molecular pathophysiology of certain types of cancer metastasis. For example, CXCR4 is a so-called tumor marker that indicates the potential for metastasis and poor prognosis in patients with breast cancer.
Small Molecule Chemokine Receptor Inhibitors.
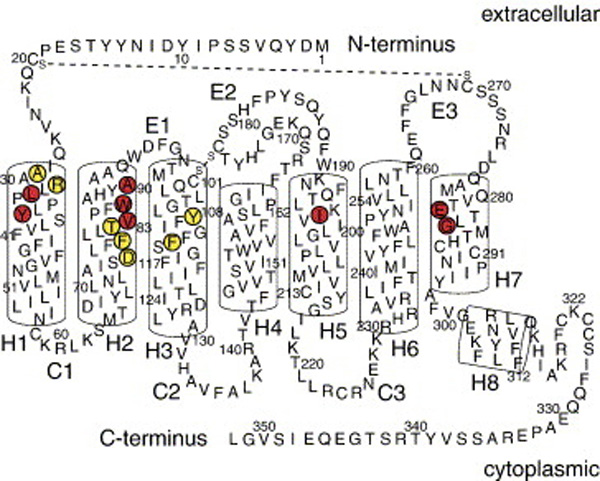
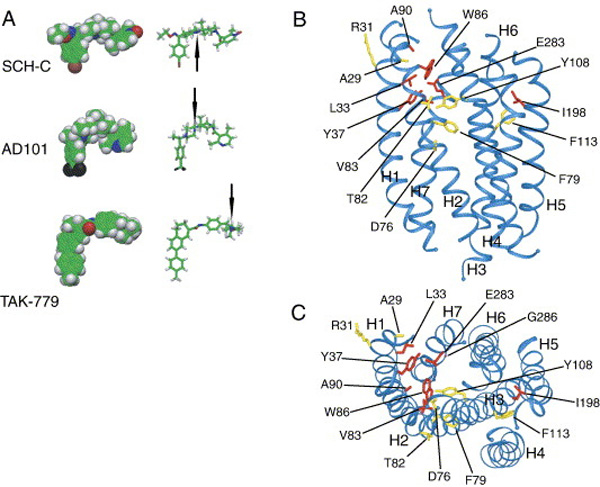
Beginning in 1998, researchers in the Sakmar laboratory together with collaborators in the laboratories of John P. Moore (Weill Medical College of Cornell University), Tatjana T. Dragic (Albert Einstein College of Medicine), and Steven O. Smith (Stony Brook University) performed extensive studies of various small molecule HIV-1 entry inhibitors that target the entry co-receptors CXCR4 and CCR5. These studies were aimed at identifying the small molecule binding site(s) of the co-receptors and at elucidating the molecular mechanism(s) underlying inhibition of HIV-1 entry by these compounds. The small molecules tested included the CXCR4 inhibitor AMD3100 and several CCR5 inhibitors such as TAK-779, AD101, and AD115. From the results of these studies it was concluded that small molecule CCR5 inhibitors bind to a common core binding site in the transmembrane domain of the receptor which is formed by transmembrane (TM) helices 1, 2, 3, and 7. These studies, which did not identify any extracellular domain residues to be involved in an interaction with the small molecule CCR5 inhibitors, it was also concluded that these compounds prevent a productive interaction of CCR5 with HIV-1 gp120 by an allosteric mechanism. This important conclusion was later supported by independent research from other laboratories, which demonstrated that small molecule CCR5 inhibitors prevent chemokine binding by a similar allosteric mechanism. Furthermore, it has been demonstrated recently that CXCR4 inhibitors such as the bicyclam AMD3100 and the monocyclam AMD3465 also bind to a small molecule binding site that is located in the transmembrane domain of the receptor. Like the CCR5-specific small molecule inhibitors, these compounds are believed to block viral entry and chemokine signaling by allosteric mechanism
Rational Drug Discovery.
The published work of the Sakmar laboratory and collaborators on small molecule CCR5 inhibitors, which has been widely cited by researchers in the HIV-1 co-receptor field, relied on a three-dimensional homology model of the transmembrane domain of CCR5 that was constructed using the 2.8 Å crystal structure of bovine rhodopsin as a template. Improved versions of this homology model, which take into account structural information from other GPCR crystal structures, that have been published more recently, are now available and will be used for mapping studies with other inhibitors. Furthermore, the work of the Sakmar laboratory and collaborators on small molecule chemokine receptor inhibitors also resulted in the generation of a large number of site-specific CCR5 and CXCR4 mutants (over 300), which will be very useful for future studies of structure-activity relationships.
Viral entry and replication assays are most relevant for the design and characterization of a potent HIV-1 inhibitor. In particular, viral entry assays using pseudotyped HIV-1 reporter viruses have been widely used by the Sakmar laboratory and collaborators to test inhibitor sensitivity of site-specific co-receptor mutants. Unfortunately, these viral assays can be difficult to quantify and are currently not amenable to high-throughput screening. Ligand binding-competition and functional signaling-inhibition assays on the other hand are commonly used by the pharmaceutical industry to screen for GPCR antagonists. For example, drug development programs at Schering-Plough, Merck, Pfizer and other major pharmaceutical companies aimed at discovering lead compounds for the development of small molecule CCR5 inhibitors were based on high-throughput chemokine binding competition assays. Therefore, to streamline the design of more potent CCR5 and CXCR4 inhibitors, the Sakmar laboratory established a functional Ca-flux assay that utilizes a state-of-the-art high-throughput drug discovery platform (FDSS6000, Hamamatsu). Depending on the specific task, various cell lines can be used to perform these Ca-flux assays. For example, in initial studies THP-1 cells (human monocytic leukemia cells; ATCC TIB-202), which express endogenous CXCR4 were used to characterize a series of discrete biguanide compounds with defined chain length (see below). Alternatively, U87.MG derived cells (human glioblastoma/astrocytoma cells; ATCC HTB-14), which lack endogenous chemokine receptors have been used by the Sakmar laboratory to assay inhibition of chemokine signaling through either wild-type or mutant chemokine receptors using stable or transient heterologous expression methods.
|
|
 Drug Development | Vision & Sensory Neuroscience | AIDS | Amyloid Disease
Drug Development | Vision & Sensory Neuroscience | AIDS | Amyloid Disease