Summary.
In the interdisciplinary environment of the Sakmar Laboratory, state-of-the-art approaches in chemical biology and proteomics are employed to study the molecular mechanism of transmembrane signaling. GPCRs are complex allosteric machines that function in the heterogeneous and dynamic environment of the cell membrane. Scores of discrete and well-characterized small molecule ligands might be available to probe any particular GPCR or GPCR family. However, to characterize engineered receptors that can only be expressed at low levels and cannot be purified to homogeneity in native membranes requires powerful and innovative analytical approaches. For example, GPCRs have the potential to be post-translationally modified by N- or O-linked glycosylation, sulfation at tyrosine residues, and phosphorylated at serine/threonine residues. They also generally contain disulfide bonds in addition to reactive cysteine residues. For example, for a particular cell-type or cell-line expressing the CC chemokine receptor CCR5, what is the degree of heterogeneity in the degree of surface expressed tyrosine sulfation at the extracellular N-terminal tail region? To answer this type of question, new microscale proteomics methods are required. Likewise, to label a GPCR at a particular location with a specific chemical or fluorescent tag is not trivial. New chemical biology approaches are required. The Sakmar Laboratory has pioneered the innovation, development and application of a number of chemical biology and proteomics methods to facilitate quantitative studies of GPCR-mediated signaling.
Proteomics of Chemokine Receptors.
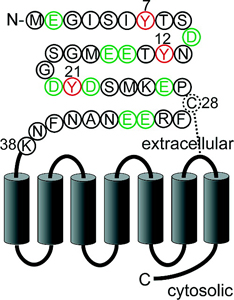
The CC-chemokine receptor 5 (CCR5), a member of the protein superfamily of GPCRs, mediates cellular responses in leukocytes induced by the CC-chemokines MIP-1a, MIP-1b, RANTES, and MCP-2. CCR5 is also the major co-receptor for the entry of macrophage trophic (R5) HIV-1 strains into target cells in vivo. In chemokine signaling and HIV-1 co-receptor function, the N-terminus of CCR5 serves as a key structure for specific ligand binding. It is known that tyrosine residues in this region are modified by sulfation and that this post-translational modification of the CCR5 N-terminus is important for high affinity interaction of the receptor with its ligands, including the HIV-1 envelope glycoprotein, gp120, in complex with CD4. Due to the lack of a well-defined substrate consensus sequence for tyrosine sulfation, it is not possible to predict which of the 4 tyrosine residues in the N-terminus of CCR5 are modified.
In collaboration with Prof. Brian T. Chait, Rockefeller University, Christoph Seibert of the Sakmar Laboratory developed methods to analyze the sulfation pattern of the N-terminus of CCR5 using a synthetic peptide that corresponds to amino acids 2 to 18 of the receptor (CCR5 2-18) as a model system. In an enzymatic in vitro sulfation assay, recombinant human tyrosylprotein sulfotransferases, TPST-1 and TPST-2, were used to modify CCR5 2-18 and the reaction products were analyzed by a combination of RP-HPLC, proteolytic degradation, and MALDI mass spectrometry. In CCR5, Tyr-14 and Tyr-15 are sulfated first, followed by Tyr-10 and Tyr-3. These results validated a method to prepare large amounts of highly sulfated peptide for structural studies, and introduced a new method to analyze complex mixtures of sulfo-peptides on a nano-scale.
The methods were extended to CXC-chemokine receptor 4 (CXCR4), a GPCR for stromal cell-derived factor-1 (SDF-1/CXCL12). SDF-1 induced CXCR4 signaling is indispensable for embryonic development and crucial for immune cell homing and has been implicated in metastasis of numerous types of cancer. CXCR4 also serves as the major co-receptor for cellular entry of T-cell line-tropic (X4) HIV-1 strains. Tyrosine residues in the N-terminal tail of CXCR4, which are post-translationally sulfated, are implicated in the high affinity binding of SDF-1 to CXCR4. However, the specific roles of three potential tyrosine sulfation sites were not well understood.
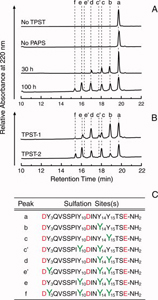
Dr. Seibert investigated the pattern and sequence of CXCR4 sulfation, again using TPST-1 and TPST-2 to modify a peptide that corresponds to amino acids 1-38 of the receptor (CXCR4 1-38). He analyzed the reaction products with a combination of reversed-phase HPLC, proteolytic cleavage, and mass spectrometry, and found that Tyr-21 is sulfated first, followed by Tyr-12 or Tyr-7. Using heteronuclear NMR spectroscopy, in collaboration with Prof. Brian F. Volkman, Medical College of Wisconsin, SDF-1 binding affinity of CXCR4 1-38 was shown to increase with the number of sulfotyrosines present. These results provided a structural basis for understanding the role of post-translational tyrosine sulfation in SDF-1-induced CXCR4 signaling, and led to the publication of the first structure of a chemokine in complex with a GPCR fragment.
New Chemical Biology Approaches to Label GPCRs.
The innovative application of recent molecular and chemical biology approaches provides an opportunity for a new dimension of quantitative structure-function studies of GPCRs. Members of the Sakmar Laboratory, in collaboration with Prof. U. L. RajBhandary, Massachusetts Institute of Technology, have recently adapted the well-known technique of amber codon suppression to expand the genetic code and allow the incorporation of several unnatural amino acids into GPCRs expressed in mammalian cells in culture. These methods were previously only useful in E. coli expression systems. In proof of concept studies, the Sakmar Laboratory incorporated the unnatural amino acids p-acetyl-L-phenylalanine (AcF), p-benzoyl-L-phenylalanine (BzF), and p-azido-L-phenylalanine (AzF), into CCR5 at high efficiency in mammalian cells to produce functional receptors harboring a single reactive keto or azido group at various positions defined by site-directed incorporation of an amber stop codon in the expressed gene sequence. Functional mutant CCR5 was obtained at levels of up to ~50% of wild-type as judged by immunoblotting, cell surface expression and ligand-dependent calcium flux assays. Rhodopsin containing AcF at different sites was also purified in high yield and reacted with fluorescein hydrazide in vitro to produce fluorescently-labeled rhodopsin.
|
|
 Membrane Biophysics | Computational Chemistry | Chemical Biology & Proteomics | Structural Biology | Nanoscience | Stem Cell Biology
Membrane Biophysics | Computational Chemistry | Chemical Biology & Proteomics | Structural Biology | Nanoscience | Stem Cell Biology
